class: title background-image: url(cover.jpg) # Re-annotating the Drosophila Testis' Transcriptome .footer[ - October 23, 2019 - @NISC ] --- # .center[Justin M. Fear] .header[ - About ] .fl.db.w-50pct[ - IRTA Fellow (NIDDK/NIH) - <i class="fas fa-dna"></i> Genomics - <i class="fas fa-project-diagram"></i> Gene Regulation - <img src="https://www.pngfind.com/pngs/m/594-5940907_file-icon-drosophila-melanogaster-svg-drosophila-cartoon-hd.png" width="6%" /> Drosophilist ] .fr.db.w-50pct[ - Contact: - <i class="fab fa-github"></i>jfear - <i class="fas fa-envelope-open-text"></i> justin.fear@nih.gov - <img src="https://avatars1.githubusercontent.com/u/13694414?s=400&u=056f5db90f080c2072519de480b976bc46521633&v=4" width="6%"/> geneticsunderground.com ] -- <hr> ### Project Code .fl.db.w-50pct[ **SRA** <i class="fab fa-github"></i>jfear/ncbi_remap ] .fr.db.w-50pct[ **PacBio** <i class="fab fa-github"></i>jfear/dmel_pacbio <i class="fab fa-github"></i>jfear/dmel_larval_pacbio ] --- layout: true .header[ - Introduction ] --- # *Drosophila melanogaster* <!-- Adults -->    <!-- larva -->   --- # Drosophila genome   --- # Testis-biased Gene Expression .fl.db.w-40pct[ Most differences between males and females are due to testis-biased expression. The testis uniquely expresses a large number of transcripts that are required for spermatogenesis. ]  .footer[ - Parisi et al. 2003. *Science* ] --- # .dn[Objectives] .w-90pct.absolute.t-3.pa-3.ba.br-5.bg-darkred.color-white[ ## Problem: Testis transcripts are under-represented in current annotations. ] .w-90pct.absolute.b-2.pa-3.ba.br-4.bg-darkgreen.color-white[ ## Objectives: 1. Use testis-specific RNA-Seq data to build a testis-specific transcriptome. 2. Use testis-specific PacBio data to build/validate a testis-specific transcriptome. ] --- layout: true .header[ - RNA-Seq Assembly ] --- # Drosophila Sequence Read Archive  -- .fs-150.fr[ | Strategy | Count | | ------------- | ------ | | RNA-Seq | 20,420 | | DNA-Seq | 8,007 | | ChIP-Seq | 5,898 | | Other | 21,629 | | **Completed** | 43,692 | ] --- # .absolute.t-0.w-100pct.center[SRA Project Overview]  --- # .absolute.t-0.w-100pct.center[Technical Metadata]   --- # Quality Control  --- # .dn[Metadata Workflow]  --- # Data Accessibility  .fl.w-40pct[ We are releasing all data to GEO. * Coverage Counts * BigWig Tracks * Sample Metadata ] --- # Testis RNA-Seq (109 samples)   --- # StringTie .w-30pct.db[Testis RNA-Seq data is used to assembly testis-specific transcriptome.]  .footer[ - Pertea et al. 2015. *Nature Biotechnology* ] --- # Testis Assembly (SRA + StringTie) .db.absolute.t-10pct.l-1[   ] .absolute.b-1.l-5-12th[ | category | count | | ------------- | ------ | | Novel Exons | 44,155 | | Novel Introns | 26,307 | | Novel Loci | 12,129 | ] --- layout: true .header[ - PacBio Validation ] --- # PacBio Samples .db.absolute.l-3-12th[ | sample | machine | # HQ Polished | | ----------------- | ------------ | ------------- | | Adult male | PacBio RS II | 12,565 | | Adult testis R1 | PacBio RS II | 12,448 | | Adult testis R2 | Sequel II | --- | | Larval testis TR1 | Sequel | 83,066 | | Larval testis TR2 | Sequel II | --- | ]   --- # Iso-Seq Workflow   --- # Adult Testis  .w-40pct[ PacBio (RS II) adult testis samples captured ~8k isoforms. ] | category | count | | ------------- | ----- | | Novel Exons | 225 | | Novel Introns | 236 | | Novel Loci | 43 | --- # .fs-90[Larval Testis]  .w-4-12th[ PacBio (Sequel) larval testis samples captured ~36k isoforms. ] | category | count | | ------------- | ----- | | Novel Exons | 2,840 | | Novel Introns | 2,242 | | Novel Loci | 707 | --- # .w-100pct.absolute.t-0.center[Similar Structures]  --- # New Structures 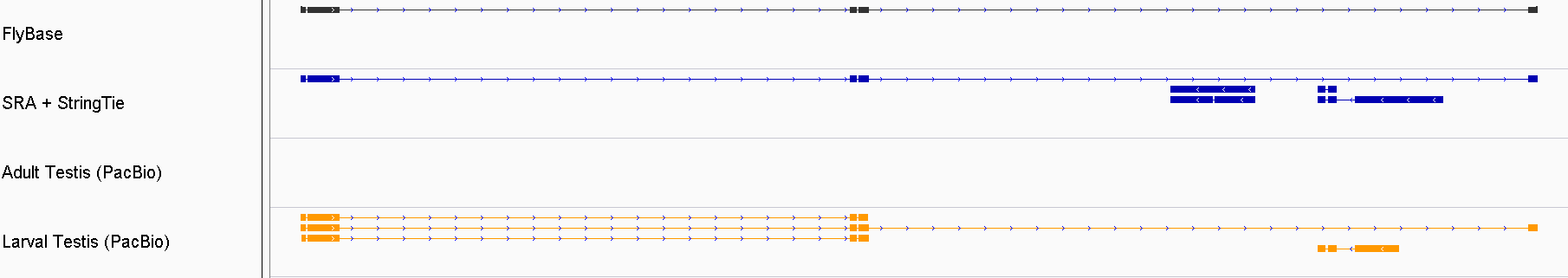  --- # .w-100pct.absolute.t-0.center[New Larval Only Structures] 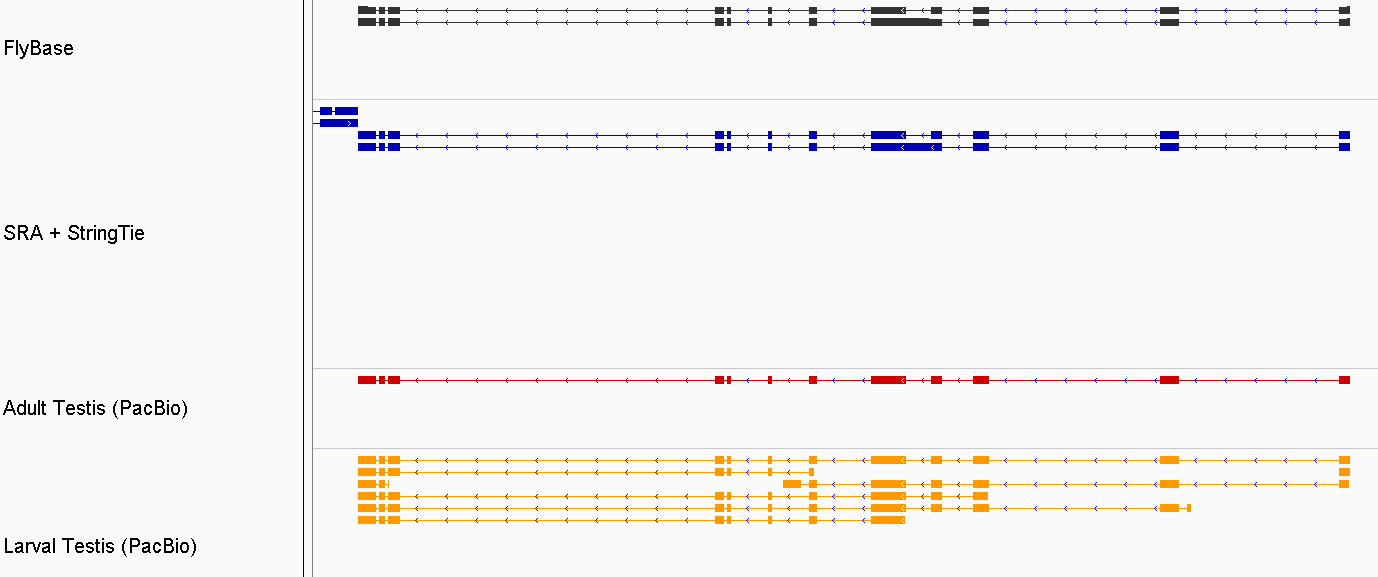 --- layout: true .header[ - justin.fear@nih.gov - geneticsunderground.com - github.com/jfear ] .footer[ - <sup>*</sup>Former Lab Members ] --- class: compact # Acknowledgements .fl.db.fs-80.mr-4[ ## Oliver Lab - Brian Oliver - Zhen-Xia Chen<sup>*</sup> - Isabelle Berger<sup>*</sup> - Sharvani Mahadevaraju - Leif Benner - Kathryn McClelland - Pradeep Bhaskar ] .fl.db.fs-80.mr-4[ ## SRA/GEO - Tanya Barrett (GEO) - Emily Clough (GEO)<sup>*</sup> - Christopher O'Sullivan (SRA) - Ben Busby (NLM) - Jean Thierry-Mieg (NLM) - Danielle Thierry-Mieg (NLM) ] .fl.db.fs-80.mr-5[ ## NISC - Alice Young - Morgan Park - Gerry Bouffard ] .fl.db.fs-80[ ## FlyBase - Norbert Perrimon - Gil dos Santos - Christopher Tabone - Lynn Crosby - Beverly Matthews - Julie Agapite - Claire Hu - Susan Russo ]